Usecase
Use-case of refTSS version 4
With the update to version 4 in refTSS, various revisions have been added, including information on candidate of cis-regulatory element (cCRE), FANTOM promoter expression table, and GWAS-LD enrichment analysis. Here, we briefly introduce use-case of the refTSS4 for the biomedical researches.
1. Surveillance of potential candidates for the promoter and proximal cis-elemment of the gene of interest.
In refTSS, it is possible to search using TSS ID, gene symbol, and gene ID from the TSS database. This result includes not only standard annotations such as gene, transcript and protein information, but also supports cCREs information for the first time in version 4. For example, promoter candidates supported by DNase hypersensitve site and histone modification have the PLS in the cCRE category, and when investigating regulatory regions, they have the cCRE categories. This provides the basis for deciding which TSS to select for study among multiple TSSs found in one gene locus.
Here we start to search the "ATF3" gene in TSS database. ATF3 is a transcription factor of the CREB/ATF family, involved in various cellular processes including inflammatory responses and the cell cycle. I plan to investigate the promoters and cis-elements that regulate this transcription factor by conducting a search.
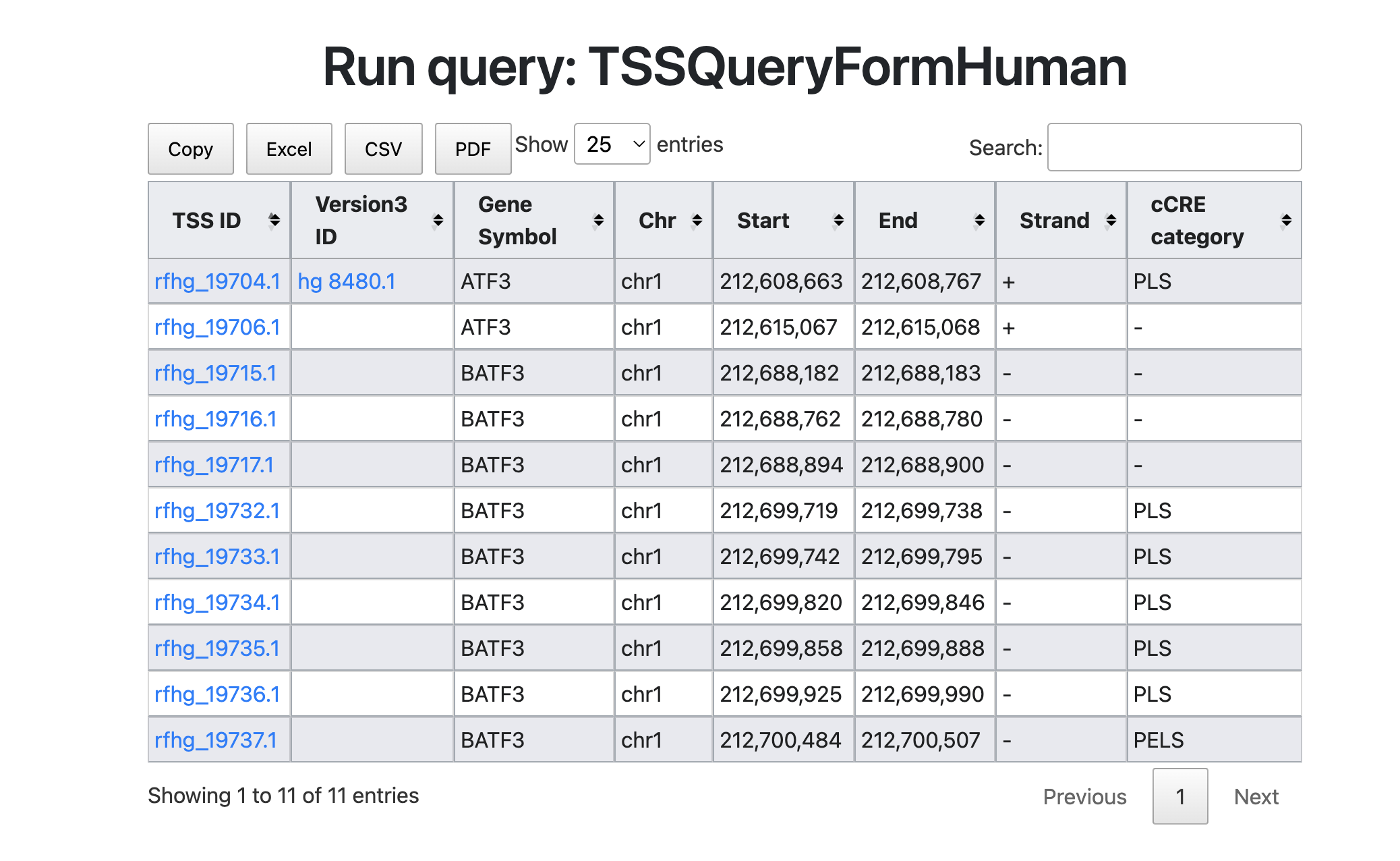
The figure above shows the search results for the keyword "ATF3." This results include not only ATF3 but also BATF3 (Basic leucine zipper transcription factor, ATF-like 3). Although BATF3 is located in a chromosomal region close to ATF3, it is a distinct gene. Therefore, two TSSs have been annotated for the ATF3 gene. BATF3 shows several TSSs overlapping with PLS and pELS. Users can survey cis-element information using refTSS-cCRE connections.
2. Investigating differences in expression levels between splicing variants using FANTOM5 TSS expression table
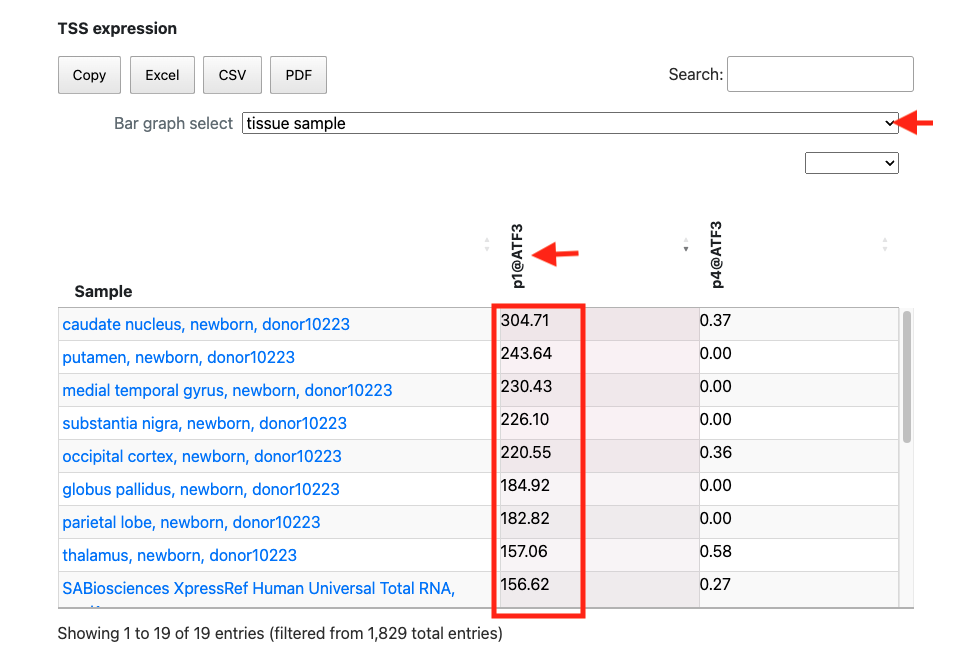
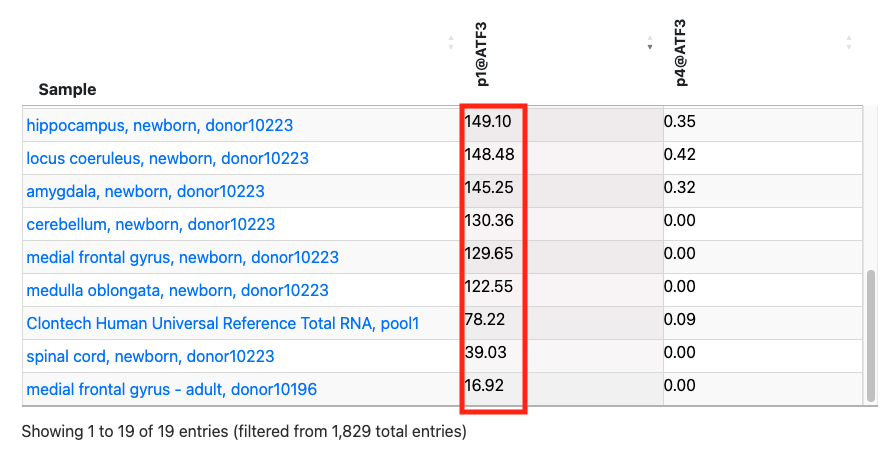
In the above figure, rfhg_19704.1 is displayed and its annotations are checked. The expression table is pickuped from at the bottom of page. Then, the FANTOM5 promoter ID "p1@ATF3" is clicked on to sort it by expression level. Additionally, by selecting the tissue sample in the Bar graph select, tissue-derived samples are extracted. While the promoter p4@ATF3 exhibits low expression in 19 tissue samples, p1@ATF3 is expressed in all tissue samples. This suggests that the main ATF3 promoter is p1@ATF3, which overlaps with TSS rfhg_19704.1.